Frequently Asked Questions¶
General Questions¶
Q: When should I use this library instead of the default 0.5 threshold?
A: You should consider optimizing thresholds when:
Your dataset is imbalanced (one class much more frequent than others)
Different types of errors have different costs (e.g., medical diagnosis, fraud detection)
You need to maximize a specific metric (F1, precision, recall) rather than general accuracy
Your model probabilities are well-calibrated but the default threshold performs poorly
Q: How does this differ from adjusting class weights in the classifier?
A: Class weights affect the model’s learning process during training, while threshold optimization is a post-training technique. They serve different purposes:
Class weights: Change how the model learns from imbalanced data
Threshold optimization: Find the best decision boundary after training
You can (and often should) use both techniques together.
Q: Can I use this with any type of classifier?
A: Yes, as long as your classifier outputs probabilities or scores. The library works with:
Scikit-learn classifiers (
predict_proba())XGBoost, LightGBM (
predict_proba()or raw scores)Neural networks (softmax outputs)
Any model that provides calibrated probability estimates
Q: What’s the difference between binary and multiclass optimization?
A:
Binary: Finds a single optimal threshold for the positive class
Multiclass: Finds per-class thresholds using One-vs-Rest strategy, then applies sophisticated decision rules for final predictions
Multiclass is automatically detected when you provide a 2D probability matrix.
Technical Questions¶
Q: Which optimization method should I use?
A: Use method='auto' (default) for most cases. It automatically selects the best algorithm:
sort_scan: O(n log n) exact optimization for piecewise metrics, guaranteed exact result
minimize: Uses scipy optimization with enhanced fallbacks
For specific needs:
- Large datasets: method='sort_scan'
- Highest precision: method='sort_scan'
- Non-piecewise metrics: method='minimize'
Q: What does “piecewise-constant” mean?
A: Piecewise-constant functions only change values at specific points (breakpoints). For classification metrics:
F1 score, accuracy, precision, recall only change when the threshold crosses a unique probability value
Between these points, the metric value stays constant
This creates “flat regions” that can trap gradient-based optimizers
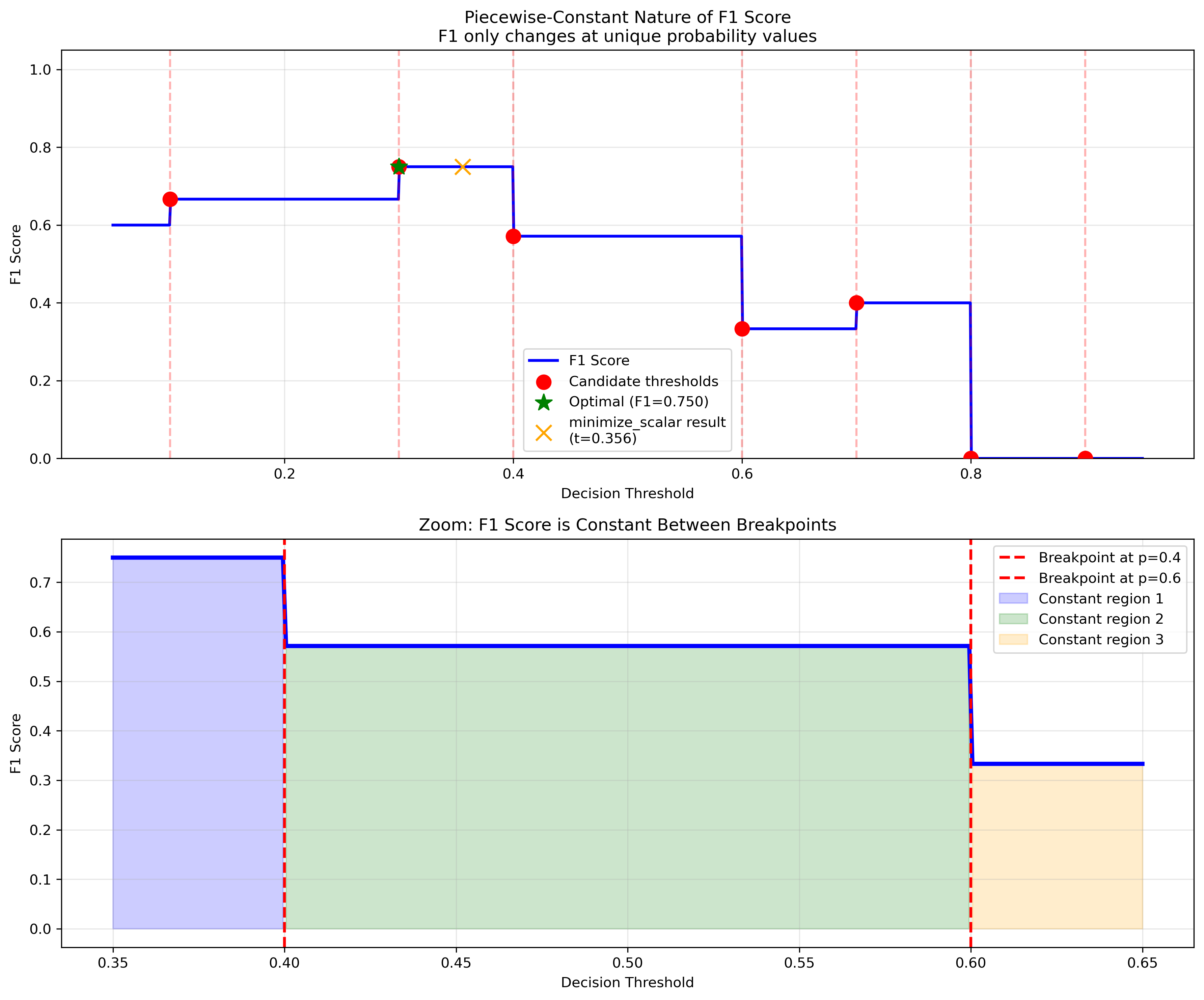
Q: How do I handle tied probabilities?
A: Use the comparison parameter:
# Exclusive: prediction = 1 if prob > threshold
result = optimize_thresholds(y, prob, comparison='>')
# Inclusive: prediction = 1 if prob >= threshold
result = optimize_thresholds(y, prob, comparison='>=')
The choice affects how samples with probabilities exactly equal to the threshold are classified.
Q: Why are my multiclass results different from argmax predictions?
A: The library uses threshold-based decision rules that can differ from simple argmax:
Apply per-class thresholds to get binary predictions for each class
Multiple classes above threshold: Predict the one with highest probability
No classes above threshold: Predict the class with highest probability
This can sometimes select a class with lower absolute probability but higher margin (probability - threshold).
Data and Performance Questions¶
Q: Do I need calibrated probabilities?
A: For best results, yes. Well-calibrated probabilities ensure that the threshold has meaningful interpretation:
from sklearn.calibration import CalibratedClassifierCV
# Apply calibration before threshold optimization
calibrated_clf = CalibratedClassifierCV(base_classifier, method='isotonic')
calibrated_clf.fit(X_train, y_train)
y_prob = calibrated_clf.predict_proba(X_test)[:, 1]
result = optimize_thresholds(y_train, y_prob_train, metric='f1')
Q: How much data do I need for reliable threshold optimization?
A: General guidelines:
Minimum: ~100 samples per class for basic optimization
Recommended: ~1000 samples per class for stable results
Cross-validation: Use CV with smaller datasets to get robust estimates
For very small datasets, consider using cross-validation or bootstrap methods.
Q: Can I use sample weights?
A: Yes, all optimization methods support sample weights:
from sklearn.utils.class_weight import compute_sample_weight
# Automatic balancing
weights = compute_sample_weight('balanced', y_train)
result = optimize_thresholds(
y_train, y_prob, metric='f1',
sample_weight=weights
)
Q: How do I handle concept drift in production?
A: Monitor threshold performance and retrain when needed:
# Regularly check if optimal threshold has drifted
current_optimal = optimize_thresholds(y_recent, y_prob_recent, metric='f1')
if abs(current_optimal - production_threshold) > 0.05:
print("Threshold drift detected - consider retraining")
See the Advanced Topics section for complete monitoring examples.
Cost-Sensitive Questions¶
Q: How do I set up cost-sensitive optimization?
A: Define utilities for each outcome type:
# Example: Medical diagnosis
medical_utilities = {
"tp": 1000, # Benefit: Early detection saves $1000
"tn": 0, # No additional cost for correct negative
"fp": -200, # Cost: Unnecessary procedure costs $200
"fn": -5000 # Cost: Missed diagnosis costs $5000
}
result = optimize_thresholds(
y_true, y_prob,
utility=medical_utilities
)
Q: What’s the difference between empirical and Bayes-optimal thresholds?
A:
Empirical: Optimizes threshold based on your training data (accounts for model miscalibration)
Bayes-optimal: Theoretical optimum for perfectly calibrated probabilities (no training data needed)
# Empirical (default)
threshold_emp = optimize_thresholds(y_true, y_prob, utility=costs)
# Bayes-optimal
result_bayes = optimize_thresholds(None, y_prob, utility=costs, mode="bayes")
Use empirical for real models; use Bayes-optimal for theoretical analysis or when you trust calibration.
Q: Can I optimize for different costs per class in multiclass problems?
A: Not directly in the current version, but you can optimize each class separately:
def multiclass_different_costs(y_true, y_prob, class_costs):
n_classes = y_prob.shape[1]
thresholds = []
for class_idx in range(n_classes):
# Convert to binary problem
y_binary = (y_true == class_idx).astype(int)
y_prob_binary = y_prob[:, class_idx]
# Use class-specific costs
costs = class_costs[class_idx]
result = optimize_thresholds(y_binary, y_prob_binary, utility=costs)
thresholds.append(result.thresholds[0])
return np.array(thresholds)
This feature is planned for future releases.
Error Messages and Troubleshooting¶
Q: I’m getting “No vectorized implementation available” - what does this mean?
A: You’re trying to use method='sort_scan' with a metric that doesn’t have a vectorized implementation:
# This will fail if 'custom_metric' lacks vectorized version
result = optimize_thresholds(y, prob, metric='custom_metric', method='sort_scan')
# Solutions:
# 1. Use different method
result = optimize_thresholds(y, prob, metric='custom_metric', method='minimize')
# 2. Register vectorized version of your metric
from optimal_cutoffs.metrics import register_metric
register_metric('custom_metric', custom_func, vectorized_func=custom_vectorized)
Q: Why am I getting “Labels must be consecutive integers starting from 0”?
A: Multiclass optimization requires class labels to be [0, 1, 2, ..., n_classes-1]:
# Bad: labels are [1, 2, 3, 4]
y_bad = np.array([1, 2, 3, 4, 1, 2])
# Good: labels are [0, 1, 2, 3]
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y_good = le.fit_transform(y_bad)
result = optimize_thresholds(y_good, y_prob, metric='f1')
Q: The optimization is very slow - how can I speed it up?
A: Several strategies:
# 1. Use faster method for large datasets
result = optimize_thresholds(y, prob, metric='f1', method='sort_scan')
# 2. Reduce data size with sampling
from sklearn.model_selection import train_test_split
y_sample, _, prob_sample, _ = train_test_split(y, prob, test_size=0.7, stratify=y)
result = optimize_thresholds(y_sample, prob_sample, metric='f1')
# 3. Use simpler comparison operator
result = optimize_thresholds(y, prob, metric='f1', comparison='>') # Slightly faster than '>='
Q: My cross-validation results have high variance - is this normal?
A: Some variance is expected, but high variance suggests:
from optimal_cutoffs.cv import cross_validate
# Check CV results
cv_results = cross_validate(y, prob, metric='f1', cv=10)
thresholds = cv_results['thresholds']
scores = cv_results['scores']
print(f"Threshold std: {np.std(thresholds):.3f}")
print(f"Score std: {np.std(scores):.3f}")
# High variance solutions:
# 1. Increase data size
# 2. Use more folds: cv=10 or cv=20
# 3. Use stratified CV for imbalanced data
# 4. Consider ensemble methods
Integration Questions¶
Q: How do I integrate this with scikit-learn pipelines?
A: Use the ThresholdOptimizer class:
from sklearn.pipeline import Pipeline
from optimal_cutoffs import ThresholdOptimizer
# Note: This requires custom pipeline components to extract probabilities
# See examples for full implementation
# Simpler approach: Apply threshold optimization after pipeline
pipeline = Pipeline([('scaler', StandardScaler()), ('clf', LogisticRegression())])
pipeline.fit(X_train, y_train)
y_prob = pipeline.predict_proba(X_train)[:, 1]
optimizer = ThresholdOptimizer(metric='f1')
optimizer.fit(y_train, y_prob)
# For predictions
y_prob_test = pipeline.predict_proba(X_test)[:, 1]
y_pred = optimizer.predict(y_prob_test)
Q: Can I save and load optimized thresholds?
A: Yes, use pickle or joblib:
import joblib
from optimal_cutoffs import ThresholdOptimizer
# Save
optimizer = ThresholdOptimizer(metric='f1')
optimizer.fit(y_train, y_prob_train)
joblib.dump(optimizer, 'threshold_optimizer.pkl')
# Load
loaded_optimizer = joblib.load('threshold_optimizer.pkl')
y_pred = loaded_optimizer.predict(y_prob_test)
Q: How do I use this with deep learning frameworks?
A: Extract probabilities from your model and apply threshold optimization:
# PyTorch example
import torch
model.eval()
with torch.no_grad():
outputs = model(X_test_tensor)
y_prob = torch.softmax(outputs, dim=1).cpu().numpy()
# For binary classification, use column 1
result = optimize_thresholds(y_test, y_prob[:, 1], metric='f1')
# TensorFlow/Keras example
y_prob = model.predict(X_test)
result = optimize_thresholds(y_test, y_prob, metric='f1')
Still Have Questions?¶
If you don’t find your question answered here:
Check the Examples for comprehensive code samples
Review the API Reference for detailed function documentation
Look at the Theory and Background section for mathematical background
Open an issue on GitHub for bug reports or feature requests
Common patterns and solutions are continuously added to this FAQ based on user feedback.